BrainStars: 1438987_at: Fam71d
| Affymetrix Probe set ID | 1438987_at |
|---|---|
| Gene Name | family with sequence similarity 71, member D |
| Gene Symbol | Fam71d GARI-L2 4921509E07Rik 4930516C23Rik |
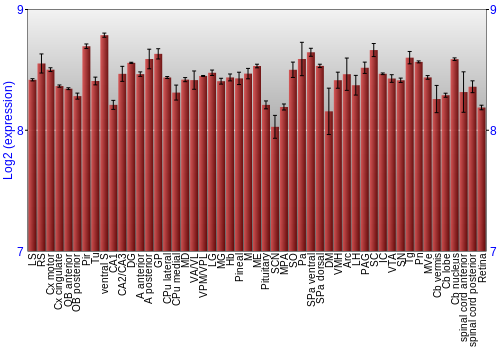
BrainStars Expression at 51 brain regions
PDF files:
Expression Graph,
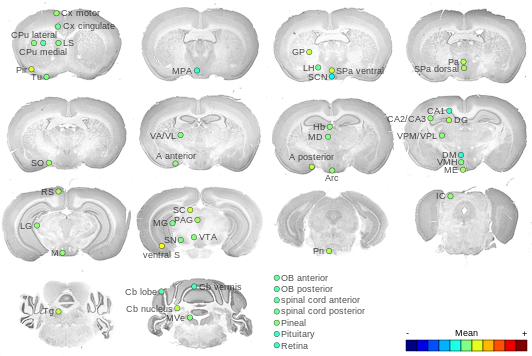
Expression Map
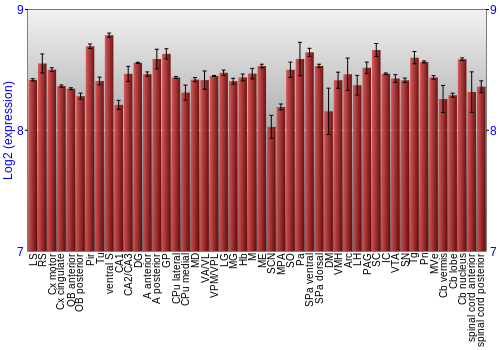
BrainStars classification of 48 brain regions by multi-state gene analysis
| # States | 1 |
|---|---|
| Score | 8.9170759134747 |
PDF files:
Expression Graph,
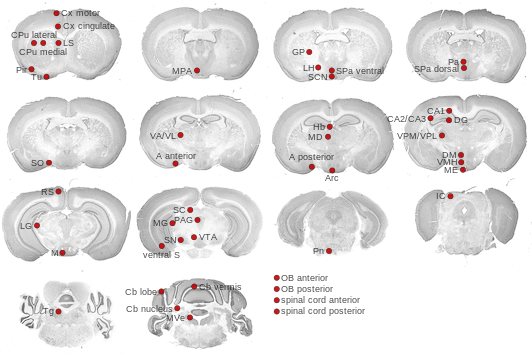
Expression Map
Other brain expression resources
Cross section images
The ViBrism cross section images (whole brain mode: beta version) were provided by the ViBrism team. A full 3-D model of the expression is available at their ViBrism site.
The ABA expression energy cross section images were obtained from the Allen Mouse Brain Atlas, Allen Instiute for Brain Science © 2008. Please refer to their original terms of use.
Links to other resources
| Allen Brain Atlas | 4921509E07Rik |
|---|---|
| ViBrism | 4921509E07Rik |
| EMAGE | Fam71d |
BrainStars 1-D expression files
You can download BrainStars expression data in a table.| TSV format file | BrainStars expression in a tab-separated values format |
| CSV format file | BrainStars expression in a camma-separated values format |
BrainStars 3-D expression files
You can download this entry's BrainStars expression data putting on 3-D mouse brain models.| VCAT format file | BrainStars expression putting on a brain model. The file can be viewed with a software, VCAT, downloadable from VCAD@RIKEN site. |
| XPR format file | BrainStars expression putting on a Allen Reference Atlas brain model. You can view the XPR file with the Brain Explorer software provided by the Allen Institute. |
External links
| Entrez GeneID | 70897 |
|---|---|
| Ensembl | ENSMUSG00000056987 |
| UniGene ID | Mm.56514 |
| RefSeq Transcript | NM_027597 NM_029069 |
| RefSeq Protein | NP_081873 NP_083345 |
| Agilent ID | A_51_P466298 |
| Allen Brain Atlas | 4921509E07Rik |
| ViBrism | 4921509E07Rik |
| EMAGE | Fam71d |